Publications
Here you can find our recent publications.
Selected

Multi‐Omics Factor Analysis—a framework for unsupervised integration of multi‐omics data sets
Molecular Systems Biology
·
18 Jun 2018
·
doi:10.15252/msb.20178124
MOFA is a factor analysis model that provides a general framework for the integration of multi-omic data sets in an unsupervised fashion
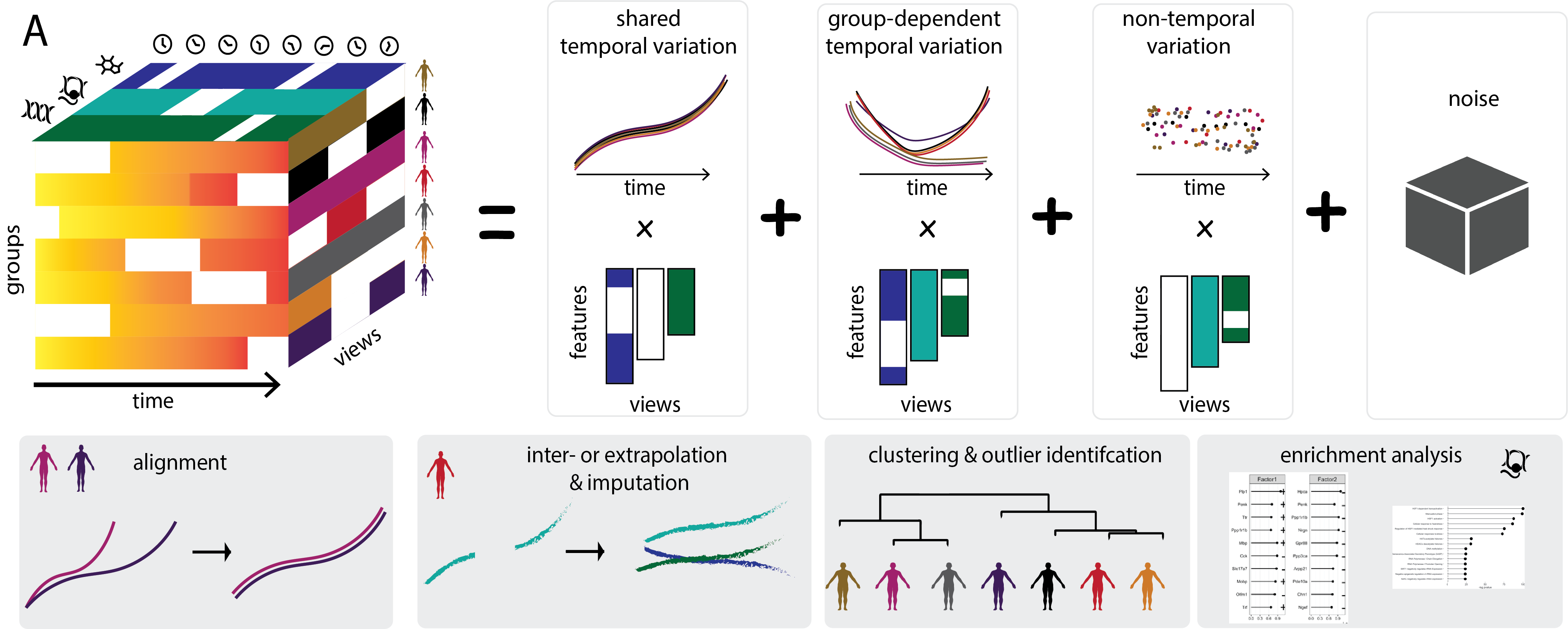
Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO
Nature Methods
·
13 Jan 2022
·
doi:10.1038/s41592-021-01343-9
MEFISTO is a unsupervised method to integrate multi-modal data with continuous structures among the samples, e.g. in spatial or temporal data.

FISHFactor: a probabilistic factor model for spatial transcriptomics data with subcellular resolution
Bioinformatics
·
11 Apr 2023
·
doi:10.1093/bioinformatics/btad183
FISHFactor identifies spatial gene expression patterns at subcellular resolution

Guide assignment in single-cell CRISPR screens using crispat
Bioinformatics
·
01 Sep 2024
·
doi:10.1093/bioinformatics/btae535
crispat is a Python framework for assigning cells to guides in pooled single cell CRISPR assays
All
2024
A genome-scale single cell CRISPRi map oftransgene regulation across human pluripotent stem cell lines
Cold Spring Harbor Laboratory
·
28 Nov 2024
·
doi:10.1101/2024.11.28.625833

Guide assignment in single-cell CRISPR screens using crispat
Bioinformatics
·
01 Sep 2024
·
doi:10.1093/bioinformatics/btae535
crispat is a Python framework for assigning cells to guides in pooled single cell CRISPR assays
In situ analysis of osmolyte mechanisms of proteome thermal stabilization
Nature Chemical Biology
·
29 Feb 2024
·
doi:10.1038/s41589-024-01568-7
2023
The changing career paths of PhDs and postdocs trained at EMBL
eLife
·
23 Nov 2023
·
doi:10.7554/elife.78706
Multicellular factor analysis of single-cell data for a tissue-centric understanding of disease
eLife
·
22 Nov 2023
·
doi:10.7554/elife.93161
Principles and challenges of modeling temporal and spatial omics data
Nature Methods
·
14 Sep 2023
·
doi:10.1038/s41592-023-01992-y

FISHFactor: a probabilistic factor model for spatial transcriptomics data with subcellular resolution
Bioinformatics
·
11 Apr 2023
·
doi:10.1093/bioinformatics/btad183
FISHFactor identifies spatial gene expression patterns at subcellular resolution
Spatial multiomics map of trophoblast development in early pregnancy
Nature
·
29 Mar 2023
·
doi:10.1038/s41586-023-05869-0
2022

Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO
Nature Methods
·
13 Jan 2022
·
doi:10.1038/s41592-021-01343-9
MEFISTO is a unsupervised method to integrate multi-modal data with continuous structures among the samples, e.g. in spatial or temporal data.
2021
Adaptive penalization in high-dimensional regression and classification with external covariates using variational Bayes.
Biostatistics (Oxford, England)
·
10 Apr 2021
·
pmid:31596468
2020
Developmental Gene Expression Differences between Humans and Mammalian Models.
Cell reports
·
27 Oct 2020
·
pmid:33113372
Survey of ex vivo drug combination effects in chronic lymphocytic leukemia reveals synergistic drug effects and genetic dependencies.
Leukemia
·
13 May 2020
·
pmid:32404973
MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data.
Genome biology
·
11 May 2020
·
pmid:32393329
Mechanistic insights into transcription factor cooperativity and its impact on protein-phenotype interactions.
Nature communications
·
08 Jan 2020
·
pmid:31913281
2019
Gene expression across mammalian organ development.
Nature
·
26 Jun 2019
·
pmid:31243369
2018

Multi‐Omics Factor Analysis—a framework for unsupervised integration of multi‐omics data sets
Molecular Systems Biology
·
18 Jun 2018
·
doi:10.15252/msb.20178124
MOFA is a factor analysis model that provides a general framework for the integration of multi-omic data sets in an unsupervised fashion
2017
Drug-perturbation-based stratification of blood cancer.
The Journal of clinical investigation
·
11 Dec 2017
·
pmid:29227286